Recently, a joint team from the College of Chemistry and Chemical Engineering, Pen-Tung Sah Institute of Micro-Nano Science and Technology, Institute of Artificial Intelligence, AI4EC Lab of the Tan Kah Kee Innovation Laboratory, Beijing AI for Science Institute, and DP Technology has achieved a significant advance in nuclear magnetic resonance (NMR) spectral analysis with the deep-learning framework NMRNet. Published in Nature Computational Science under the title “Toward a unified benchmark and framework for deep learning-based prediction of nuclear magnetic resonance chemical shifts,” the work introduces an innovative SE(3) Transformer architecture that delivers high-accuracy chemical shifts predictions for both solution-state and solid-state NMR, providing a powerful new tool for molecular structure elucidation and materials design.
As deep learning techniques have matured, researchers have increasingly explored their application to NMR chemical shifts prediction, aiming to enhance model accuracy and efficiency. Deep learning approaches—particularly graph convolutional networks (GCNs) and equivariant message-passing neural networks (MPNNs)—have already demonstrated superior precision over traditional methods in the context of solution-state NMR and are able to handle the complexity of molecular structures with remarkable fidelity. Nonetheless, such solution-state models typically disregard intermolecular interactions, especially solvent–solute effects, although recent efforts have sought to compensate by integrating molecular dynamics simulations and first principles calculations. In solid-state NMR, accounting for periodic boundary conditions (PBC) is indispensable. Here, machine learning models trained on density functional theory (DFT) data have begun to deliver both high efficiency and high accuracy. Yet existing models are overwhelmingly specialized for either solution or solid states, and their general applicability remains insufficiently validated, leaving notable limitations unaddressed.
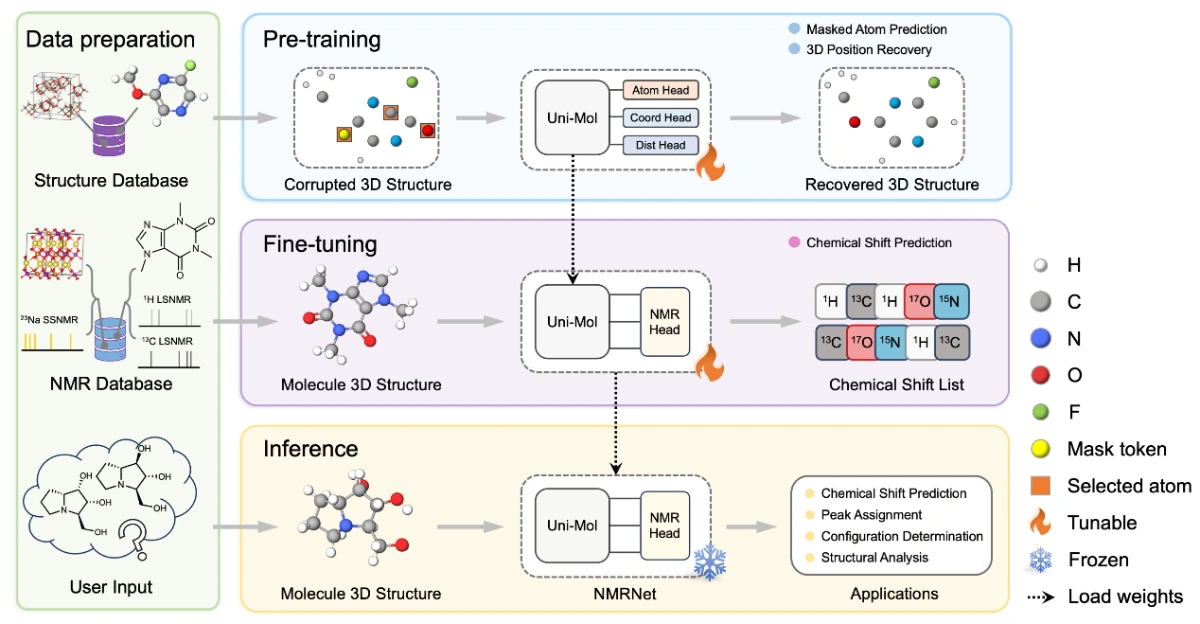
The College of Chemistry and Chemical Engineering, Pen-Tung Sah Institute of Micro-Nano Science and Technology, Institute of Artificial Intelligence, AI4EC Lab of the Tan Kah Kee Innovation Laboratory, Beijing AI for Science Institute, and DP Technology jointly developed the NMRNet framework. By adapting the SE(3) Transformer architecture from Uni-Mol within a pretraining–fine-tuning paradigm, NMRNet enables unified modeling of liquid, solid, and gaseous systems. The team curated structural and NMR data, extracted 3D information, and established a standardized benchmark dataset, nmrshiftdb2-2024, after extensive data cleaning and validation. Pretraining reused Uni-Mol weights for liquid NMR and performed self-supervised learning on over 4.8 million crystal structures for solid NMR, effectively addressing data scarcity. Fine-tuning across multiple datasets demonstrated state-of-the-art accuracy in both single- and multi-element prediction. NMRNet further supports peak assignment and conformation analysis, advancing the study of structure–spectrum relationships.
The first author of this work is Fanjie Xu, a master’s student at the College of Chemistry and Chemical Engineering, Xiamen University. The corresponding authors are Prof. Jun Cheng, Prof. Fujie Tang, and Zhifeng Gao (Research Scientist, DP Technology). Collaborators include Dr. Feng Wang (Associate Research Fellow, AI4EC Lab) as well as Lin Yao and Hongshuai Wang (Research Scientists, DP Technology). The study benefited from the guidance of Academicians Zhong-Qun Tian and Weinan E, and received support from Dr. Linfeng Zhang, Director of the Beijing AI for Science Institute.We thank Y. Ren and J. Zhang for their contributions to the design of the manuscript’s cover. We thank Y. Tang and J. Qiu for his valuable improvements to the schematic diagram. We are also grateful for the insightful discussions and suggestions from Y. Liu, J. Zou, Y. Zhuang, Y. Jin, F. Fu, W. Luo, G. Zhou and J. Wang. F.T. acknowledges the National Key R&D Program of China (grant no. 2024YFA1210804) and a startup fund at Xiamen University. J.C. acknowledges the National Natural Science Foundation of China (grant nos. 22225302, 92470201, 22021001, 92461312, 21991151, 21991150, 92161113 and 22411560277), the Fundamental Research Funds for the Central Universities (20720220009), Laboratory of AI for Electrochemistry (AI4EC), IKKEM (grant nos. RD2023100101 and RD2022070501).
DOI:
https://doi.org/10.1038/s43588-025-00783-z
Github:
https://github.com/Colin-Jay/NMRNet
Data set:
https://zenodo.org/records/13317524
APP:
https://ai4ec.ac.cn/apps/nmrnet
https://bohrium.dp.tech/apps/nmrnet001
